Tissue Distribution and Clonal Diversity
Tissue Distribution and Clonal Diversity
Autoreactive CD4+ and CD8+ T cells have been identified from the peripheral blood and islets of type 1 diabetes (T1D) patients and organ donors, and autoantibodies secreted by autoreactive B cells are the most widely used biomarker of T1D. However, the T1D adaptive immune repertoire had not been examined without preselecting for known cell reactivities. Moreover, questions exist regarding the degree of repertoire overlap across tissues, across T cell subsets, whether specific clones are common across individuals with T1D, and whether analyses of the adaptive immune repertoire could be used to distinguish T1D from control subjects. Seay et al. examined the T cell receptor (TCR) β chain (TRB) and B cell receptor (BCR) immunoglobulin heavy chain (IGH) repertoire by high throughput immunosequencing. Samples were compared from T1D and control organ donor tissues including peripheral blood, pancreatic draining lymph node (pLN), “irrelevant” non-pancreatic draining lymph node (ILN), spleen, and for one T1D donor, the intra-islet T cells of the insulitis lesion. First, cells were sorted to identify CD4+ conventional T cells (Tconv), CD4+ regulatory T cells (Treg), CD8+ T cells, and B cells. From each sorted cell subset, the authors sequenced genomic DNA encoding the highly polymorphic complementarity determining region 3 (CDR3) spanning the V(D)J junction of the TCR or BCR, which is responsible for antigen engagement. The sequences were subjected to bioinformatics analyses to determine TCR and BCR gene family member usage, V-J gene pairing, repertoire diversity, sequence overlap across tissues (for each individual donor), and sequence overlap across donors. The corresponding amino acid sequences were determined and compared against known autoreactive T cell clones from the literature, to determine probable reactivity. The library of sequence data generated is publicly available at http://clonesearch.jdrfnpod.org/.
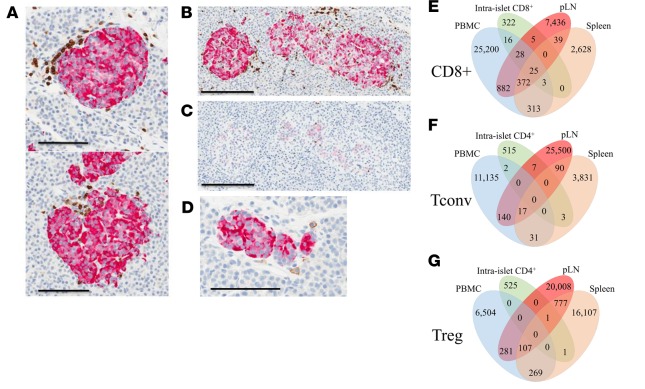
Overall, the authors found that TCR and BCR V-J gene usage and pairing as well as receptor diversity were comparable in T1D versus control donors. While CD8+ TCR sharing was equally common in HLA-class I matched T1D and control donors, there were 14 unique CDR3 amino acid sequences shared across all seven T1D donors carrying HLA-A*02. These CDR3 sequences did not correspond to clones previously reported in the literature, so their antigen reactivities and activation states remain unknown at this time. The authors reported that the Tconv, Treg, and B cell repertoires were highly tissue restricted with little clonal overlap across tissues. In contrast, overlap across was more common for CD8+ T cells, suggesting this subset to offer the greatest potential as T1D biomarkers measurable from peripheral blood in living patients. Surprisingly, these observations were true even when comparing the islet and pLN repertoire from a single donor; however, the anatomical position of the pLN in relation to the pancreas was not known. It remains unknown whether CD4+ T cell clonal overlap may be more common in the pLN nearest to an inflamed islet. 48 CDR3 amino acid sequences identified from the various tissues and T cell subsets corresponded to previously described autoreactive TCR clones, and 24 of them were significantly more frequent in T1D donors versus donors without T1D. Importantly, the CDR3 amino acid sequence corresponding to a known GAD-reactive TCR clone (4.13) was highly enriched in tissues from seven T1D donors, including the intra islet sample, and one control with T1D-permissive HLA, though the clone’s potential pathogenic significance remains to be explored.
The T cell repertoire develops within the context of HLA, restricting the ability to make comparisons across individuals carrying different HLA alleles. Because large cohorts of HLA-matched subjects were not available within the available nPOD biorepository, the current study was unable to definitively determine an adaptive immune repertoire signature for T1D. However, this study represents a first in man effort of its kind that lays the groundwork for efforts evaluating the potential for T cells to serve as biomarkers of T1D to guide future research efforts and potentially, clinical decisions. Proof of principle studies (by Emerson et al.) of the adaptive immune repertoire confirmed that immunosequencing can accurately discriminate subjects who are seropositive for cytomegalovirus (CMV) versus those who are seronegative; hence, it seems feasible that given sufficient sample size with HLA-matching, it will be possible to determine common autoreactivities comprising a T1D adaptive immune signature to aid in predicting symptomatic disease onset in high-risk subjects. Furthermore, the findings reported by Seay et al. highlight the notion that immunosequencing data could be incredibly useful for identifying candidates for a given intervention, characterizing response to treatment, guiding go/no-go therapeutic decisions, and ultimately, developing tailored therapies for patients with or at risk for symptomatic T1D.
Tissue Distribution and Clonal Diversity of the T and B Cell Repertoire in Type 1 Diabetes.
H Seay, E Yusko, S Rothweiler, L Zhang, A Posgai, M Campbell-Thompson, M Vignali, R Emerson, J Kaddis, D Ko, M Nakayama, M Smit, J Cambier, A Pugliese, M Atkinson, H Robins, T Brusko (2016).